Laboratory : Plant Circadian Rhythm Laboratory
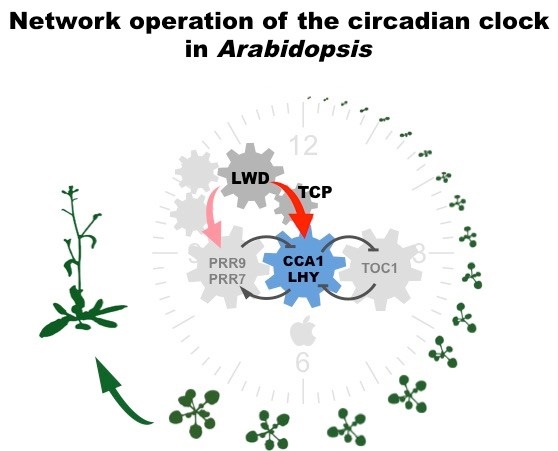
Most of organisms on the earth evolve circadian clocks to control the oscillations of multiple physiological pathways in sync with the ~24-h period, upon the day-night cycles resulted from the earth rotation, such oscillations are called “circadian rhythms”. Besides the predictable day-night and seasonal cycles, plants are sessile in circumstances with numerous unpredictable transient changes during their growth and development. The rhythmicity of the “circadian clock” profoundly keeps plants on the track without over reacting to the random cues, therefore sustains propagating on an optimal season time. The circadian clock comprises genes tightly interlocked in regulatory feedback loops, in other words, clock related genes are directly or indirectly regulated by their downstream genes. Such regulatory feedback loops make genes peak at a specific time of the day and oscillate according to the external cues like light and temperature through the day-night cycles. In the model plant, Arabidopsis , LIGHT-REGULATED WD1 (LWD1) plays a role in the morning activation of circadian clock. The five WD repeats of LWD1 form a propeller structure and serve as a protein-protein interaction platform. In our previous study, we have revealed that LWDs form co-activator protein complexes with specific TCP members to directly activate the expression of morning gene CCA1 , thus, secure the normal operation of the circadian clock at dawn. However, expressions of several clock genes in the lwd1 lwd2 double mutant don't obey the interlocking gene circuits, e.g. genes and their repressors are down regulated simultaneously when lacking LWDs. This indicates that our knowledge of the circadian clock is not thorough. To investigate the regulatory mechanism of circadian clock, we will use LWD1 as bait to identify transcription factors with similar transcription trends through a plenty of transcriptomes. We will reveal the molecular mechanism of the circadian clock operation via the study of the LWD1 co-expressed transcription factors. Our study would unravel multifaceted connections between circadian clock and plant developmental processes.
Publications
1. Meng-Chun Lin, Huang-Lung Tsai, Sim-Lin Lim, Shih-Tong Jeng, and Shu-Hsing Wu (2017) Unraveling multifaceted contributions of small regulatory RNAs to photomorphogenic development in Arabidopsis. BMC Genomics 18: 559. (5-yr Impact factor: 4.284; Rank: 33/158 in Biotechnology & applied microbiology)
2. Jing-Fen Wu*, Huang-Lung Tsai*, Ignasius. Joanito, Yi-Chen Wu, Yi-Hang Li, Chin-Wen Chang, Jong-Chan Hong, Jhih-Wei Chu, Chao-Ping Hsu and S. H. Wu (2016) LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nature Communications 7:13181 (*co-first authors)(5-yr Impact factor: 13.092; Rank: 3/64 in multidisciplinary sciences)
3. Huang-Lung Tsai, Yi-Hang Li, Wen-Ping Hsieh, Meng-Chun Lin, Ji Hoon Ahn, and Shu-Hsing Wu (2014) HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. The Plant Cell 26 (7): 2858-2872. (5-yr Impact factor: 9.975; Rank: 6/211 in Plant Sciences)(The work was selected as one of the significant publications in Academia Sinica 2014).
4. Ming-Jung Liu, Szu-Hsien Wu, Jing-Fen Wu, Wen-Dar Lin, Yi-Chen Wu, Tsung-Ying Tsai, Huang-Lung Tsai and Shu-Hsing Wu (2013) Translational landscape of photomorphogenic Arabidopsis. The Plant Cell 25 (10): 3699-3710. (5-yr Impact factor: 9.975; Rank: 6/211 in Plant Sciences)
5. Ya-Ling Lin, Shu-Chiun Sung, Hwang-Long Tsai, Ting-Ting Yu, Ramalingam Radjacommare, Raju Usharani, Antony S. Fatimababy, Hsia-Yin Lin, Ya-Ying Wang, and Hongyong Fu (2011) The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. The Plant Cell 23 (7): 2754-73. (5-yr Impact factor: 9.975; Rank: 6/211 in Plant Sciences)
6. Antony S. Fatimababy, Ya-Ling Lin, Raju Usharani, Ramalingam Radjacommare, Hsing-Ting Wang, Hwang-Long Tsai, Yenfen Lee and Hongyong Fu (2010) Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. FEBS J. 277 (3): 796-816. (5-yr Impact factor 4.129; Rank: 82/286 in Biochemistry and Molecular Biology)
7. Huang-Lung Tsai, Wei-Ling Lue, Kuan-Jen Lu, Ming-Hsiun Hsieh, Shue-Mei Wang, and Jychian Chen (2009) Starch synthesis in Arabidopsis is achieved by spatial cotranscription of core starch metabolism genes. Plant Physiology 151 (3): 1582-1595. (5-yr Impact Factor: 7.428; Rank: 11/211 in Plant Sciences)